Multimodal Contrastive Learning Of Medical Time Series And Notes
Advancing ICU Care: Multimodal Pretraining of Medical Time Series and Notes
By Ryan King, Tianbao Yang, and Bobak J. Mortazavi
Texas A&M University
Modern intensive care units (ICUs) generate vast amounts of patient data—ranging from clinical measurements like vital signs and lab results to textual clinical notes detailing physicians’ observations and treatment plans. Effectively analyzing this data is critical for advancing patient care but remains a challenging task due to data complexity, missing values, and the high cost of annotating ICU data.
Our work, presented at Machine Learning for Health (ML4H) 2023, introduces a novel multimodal pretraining approach to tackle these challenges. By aligning clinical time series and notes using self-supervised learning, we demonstrate significant advancements in downstream tasks like in-hospital mortality prediction and phenotyping.
Challenges in ICU Data Analysis
- Data Diversity and Noise: ICU data varies widely in format (e.g., numeric vs. textual) and quality, often containing noise and missing values.
- Annotation Cost: Labeling ICU data is expensive and labor-intensive, requiring domain expertise.
- Underutilized Modalities: Existing methods often focus on a single data type, neglecting the complementary nature of clinical notes and measurements.
Our Approach: Multimodal Pretraining
We propose a self-supervised multimodal pretraining framework that simultaneously aligns and reconstructs clinical measurements and notes. Key components include:
1. Alignment Pretraining with Contrastive Learning
We align measurement and text representations by maximizing the similarity between embeddings from the same ICU stay while minimizing similarity with embeddings from other stays. This ensures the two modalities are mapped into a shared semantic space.
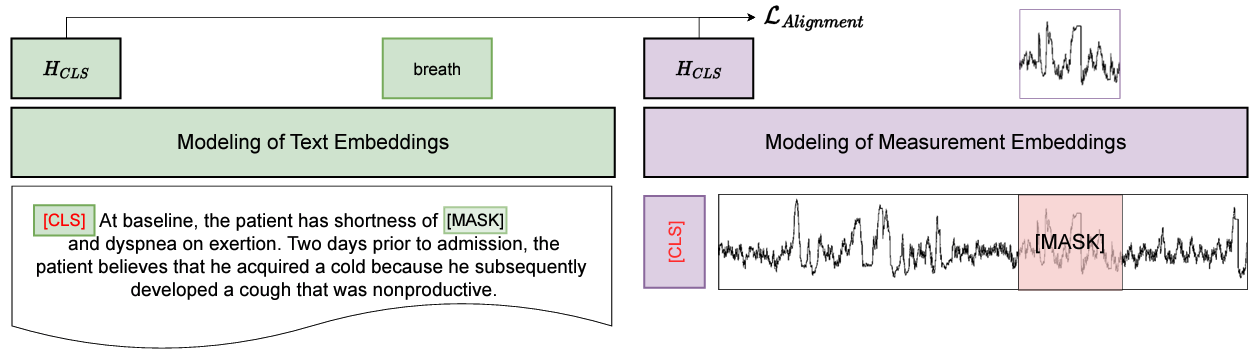
2. Masked Pretraining for Reconstruction
Inspired by models like BERT, we randomly mask tokens in clinical notes and measurement values, challenging the model to reconstruct the missing information. This enhances its ability to learn meaningful representations from incomplete data.
Experimental Results
We evaluated our model on the MIMIC-III dataset, a benchmark dataset for ICU prediction tasks. Key results include:
In-Hospital Mortality Prediction
- Our pretrained model improved AUC-ROC by 0.17 and AUC-PR by 0.1 when only 1% of labeled data was available.
- The model achieved near state-of-the-art performance even in semi-supervised settings, outperforming baselines.
Phenotyping
- For multi-class, multi-label classification of 25 phenotypes, our approach demonstrated improved micro and macro AUC scores, particularly with limited labeled data.
Zero-Shot Evaluation
Without fine-tuning, our model achieved an AUC-ROC of 0.709 and AUC-PR of 0.214 for mortality prediction—a promising step toward deploying pretrained models in real-world settings.
Why It Matters
By leveraging both clinical notes and measurements, our method captures richer patient representations, making it more adaptable to diverse ICU scenarios. The ability to perform well with minimal labeled data reduces the burden of data annotation, paving the way for scalable deployment in healthcare systems.
Limitations and Future Work
While effective, our approach has limitations:
- The expressiveness of the text encoder is constrained during alignment.
- Performance on some phenotyping tasks plateaued with higher percentages of labeled data, potentially due to catastrophic forgetting.
Future research could explore simultaneous optimization of both encoders and evaluate additional zero-shot tasks across other datasets.
Open Source Code
Our implementation is available on GitHub:
Multimodal Clinical Pretraining
The paper can be found here:
arXiv
This work represents a significant step toward making ICU data more actionable, reducing annotation costs, and improving critical care outcomes. If you’re interested in multimodal learning or healthcare AI, we’d love to hear your thoughts and collaborate on future research!
