New Student Resources
This page was created to help you get setup with the resources we have here in our lab. If you have any other information you would like to know, please send me a message and let me know.
- Server Access
- Connect to the Server
- Environment Setup
- Install Jupyter Lab
- Server Etiquette
- Useful Info
Server Access
To access the server, you first need an account on the server. Accounts are created by Engineering IT (engrit@tamu.edu). Either Dr Mortazavi or the graduate student who is advising you will email them about access. What they need from you is your netid (not your UIN). Once they have it, they’ll email IT about generating an account for you. This may take a week to do so please be patient. However, if you haven’t heard back in a week about your account, ask what the status is.
Connect to the Server
Once you have an account, you will be able to connect to the server using SSH. An example is given below:
ssh -o ServerAliveInterval=5 <netid>@cse-stmi-s1.cse.tamu.edu
Don’t worry about the ServerAliveInterval option. This just keeps your session from timing out.
Note: if you aren’t at Texas A&M you will need to install the Cisco AnyConnect VPN for your OS and connect to it before you’ll be able to connect to the STMI server. Below are the links to install the Cisco AnyConnect per OS:
Environment Setup
Now that you’re able to connect to the server, you need to set up your conda environment. To create the environment, you can run the following command:
conda create --name <env name of your choice> python=<python version>
Conda is then going to show you a list of packages it needs to install. Type ‘y’ and proceed with the creation of the environment.
Once you’ve created the environment, you can activate it using the following command:
conda activate <your env name>
For a cheat sheet of useful conda commands, click this link
Install Jupyter Lab
This part is completely optional but it’ll give you a nice clean way of viewing your code and files on the server. If you have a different method of accomplishing the same thing, go for it.
To start, install Jupyter Lab with the following command:
conda install -c conda-forge jupyterlab
Just like your environment creation, conda is going to find a list of packages it needs to install and ask if it’s ok. Type ‘y’ to proceed with the installation.
To connect to your jupyter notebook remotely, you’ll need to have jupyter lab setup on your local machine. You can do this by installing conda on your local machine and following the same process above on your local machine or you can install it without anaconda.
Once you have jupyter lab setup on your machine and the server, we’re ready to connect the two. To connect, you need a terminal open that is connected to the server and has your conda environment activated and an additional terminal to run jupyter lab locally. Then follow these steps:
- On the server, run
jupyter lab --no-browser --port=8889- You probably won’t get port 8889 because it’s busy but jupyter will find an available port and display it on your screen as a URL that starts with localhost.
- Example output is below. The higlighted portion is the link you’ll use. Notice that here it does say 8889:
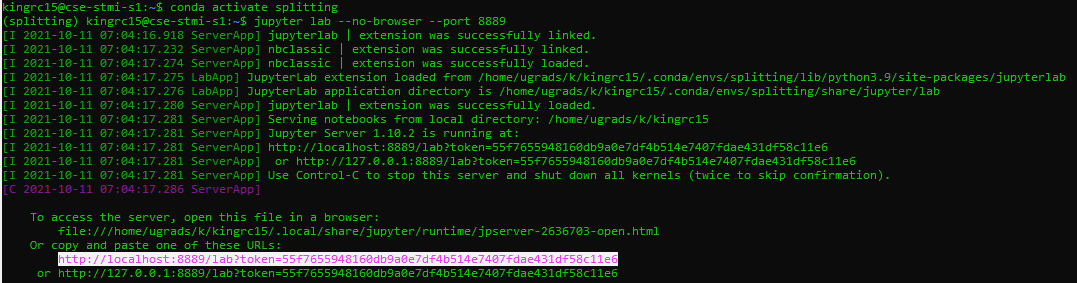
- On your local machine, run
ssh -L localhost:<port number>:localhost:<port number> <netid>@cse-stmi-s1.cse.tamu.edu- The port number is the one that was specified in step 1. Don’t forget to use your netid when connecting to the server.
- As long as you don’t have any errors, you can copy the URL provided in step 1 over to a new tab in a browser of your choice. At this point, you should be connected and able to interact with the server through the link.
Server Etiquette
renice
If you know that you’re going to be running a large task (i.e. training a model on ImageNet), change the priority of your code so that it doesn’t prevent others from running their tests. You can do this by “renicing” your code. You can use renice to change the priority of all of your jobs with this command:
renice -n <priority> -u <your username>
Priorities are set to 0 by default. Essential OS jobs will have a negative priority. If you need to change the priority your code, change it to a high positive number (i.e. 10). If you need to change the priority of a specific job, you can run this:
renice -p <pid>
You can find the PID of your jobs and monitor their resource usage with htop:
htop -u <username>
For more information about htop or renice, you can use -h to get a short description of the arguments.
gpustat
Additionally, monitor your GPU usage. The GPUs are there to be used. However, you shouldn’t be using all the GPUs all the time. To monitor your GPU usage, install gpustat:
conda install -c conda-forge gpustat
You can run gpustat from your command line using the following command:
gpustat -pi
The p flag will display the processes being run on each GPU. The i flag will launch gpustat in interactive mode. If you just want a snapshot of what the GPUs currently are doing, leave the i flag out. I recommend running this command with the i flag and placing it somewhere you can see all the time that way you can monitor your GPU usage.
Useful Info
Running code while disconnected
You may want to run your code while you aren’t connected to the server. For example, you may want something to run over night but can’t keep your SSH session open. There are two programs that can be used to run your code in these scenarious: tmux and screen. These two programs work very similarly. They both create a terminal that can be detached or reattached as needed. To create a session, just type the name of the program you’d like to run (tmux or screen). The new terminal will work exactly the same as your normal terminal except now, when you disconnect from your SSH session, your code will still be running.
Reconnecting is slightly different for tmux and screen. For both, you’ll first need to reconnect to the server. Then you’ll do the following to reconnect:
-
For
tmux, the command istmux attach. This will automatically attach you to a detached tmux session. -
For
screen, the command isscreen -r. If you have multiple screen sessions, you’ll need to specify the process idea like below:screen -r <pid>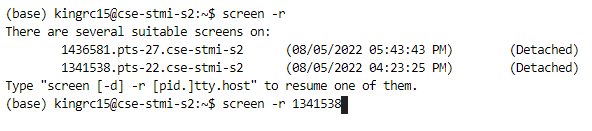
For more information about tmux, checkout this cheatsheet
For more information about screen, checkout this cheatsheet
Experiment Tracking (Weights and Biases)
If you haven’t heard of Weights and Biases, you need to check them out: https://wandb.ai/. They provide a way to track your experiments, visualize their performance and even perform hyperparameter optimization. It really simple to setup too. Just import the python library wandb, create the session and log important metrics to wandb. Weights and Biases can run your hyperparamter optimization if you use the argparse library to configure your model and training. It’ll even provide a guess a what it thinks your search space should look like when you initialize the sweep.
